SAMPLE PhD RESEARCH PROJECT IN YUE GROUP OF CSL LAB:
GENETICALLY ENCODED Ca2+ SENSORS
Fluctuations in the concentration of intracellular Ca2+ ions serve as a universal language of life, as lived out at the molecular/cellular level (e.g., in neurons and cardiac myocytes). As such, a major endeavor of modern engineering biology is to understand how this ‘microscopic language’ is produced and decoded by molecules, with the ultimate aim of manipulating such communication towards therapeutic benefit. A key step in this enterprise is to measure intracellular Ca2+ concentration in live organisms, in specific cells, and in specific locations within cells. The standard means of making such measurements is to fill cells with inorganic fluorescent molecules, whose fluorescence properties change with the level of intracellular Ca2+. The serious limitation of this approach is that long-term loading of such inorganic dyes is impractical and/or toxic, and it is very difficult to load dye into selected cell types or locations within cells.
To overcome these restrictions, we are utilizing genetically encoded Ca2+ sensors—proteins made by cells themselves, after introduction of a gene that encodes such sensors. Being proteins, these genetically encoded sensors can be innocuously deployed within live cells for extended periods of time, and genetic engineering can make it so that sensors only express in certain cell types, and in certain subcellular locations. Still, genetically encoded Ca2+ sensors face critical obstacles before going ‘prime time.’ One of the main challenges is that these sensors respond to Ca2+ fluctuations with a complex delay; in other words, these sensors are kinetically slow.
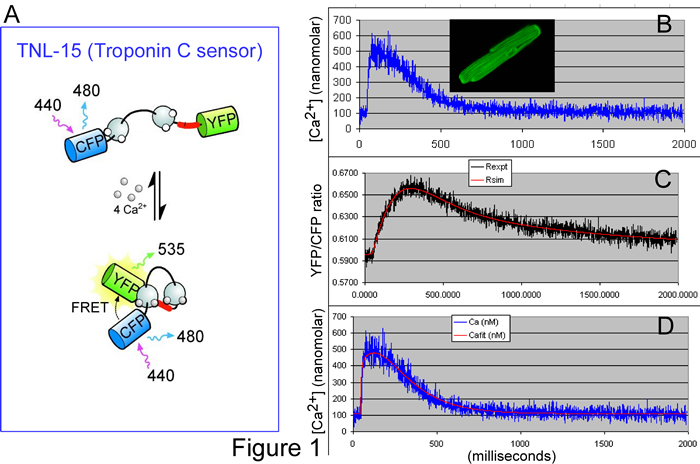
Accordingly, the BME PhD research of Lai Hock Tay seeks to determine and test a ‘transfer function’ that describes the kinetic response of TNL-15 (a leading genetically encoded Ca2+ sensor) to Ca2+ fluctuations. Figure 1A illustrates the principle of TNL-15, which is based on the naturally occurring molecule, troponin C. Normally, this molecule binds Ca2+, undergoes a conformational change, and then activates skeletal muscle contraction. In TNL-15, the ends of troponin C have been fused to two color-mutants of green fluorescent protein (GFP): namely, cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP). Based upon a physical phenomena known as fluorescence resonance energy transfer (FRET), the fluorescence of the sensor will change depending upon the level of Ca2+, as follows. When short blue light excites TNL-15, the resulting fluorescence is more cyan or yellow, depending upon the distance between CFP and YFP fluorophores. In low Ca2+, the conformation of troponin C is such that the spacing between CFP and YFP is relatively large; hence, the fluorescence ratio of YFP/CFP is relatively low. In high Ca2+, troponin C changes conformation, bringing CFP and YFP closer together, thereby increasing the YFP/CFP fluorescence ratio. TNL-15 thus monitors intracellular Ca2+ fluctuations, with the fluorescence ratio of YFP/CFP as a readout.
To determine the transfer function of TNL-15, single cardiac myocytes were loaded with a traditional inorganic Ca2+ dye (Indo 1), and also made to express TNL-15 (Figure 1B, inset). Signals from Indo 1 provide a measure of the true Ca2+ waveform (Figure 1B, blue noisy trace). Simultaneously, the YFP/CFP fluorescence ratio provides the kinetically slowed readout from TNL-15 (Figure 1C, black noisy trace). Using data such as these from many cardiac cells, we deduced the ‘forward transfer function’ for TNL-15: when the blue noisy trace from Figure 1B was fed through this transfer function, the output was the smooth red curve in Figure 1C (an impressive prediction). Of greater utility, the transfer function could be manipulated into a ‘reverse transfer function.’ If the red smooth trace in Figure 1C is input into the ‘reverse function,’ the output is the smooth red curve in Figure 1D, which provides a good prediction of the ‘true’ Ca2+ waveform (blue noisy trace in Figure 1D is reproduced from that in Figure 1B). Hence, with the aid of these transfer functions, one can backcalculate the kinetically faithful Ca2+ transient, based only on the slow readout from TNL-15, thereby overcoming one of the challenges facing genetically encoded Ca2+ sensors.
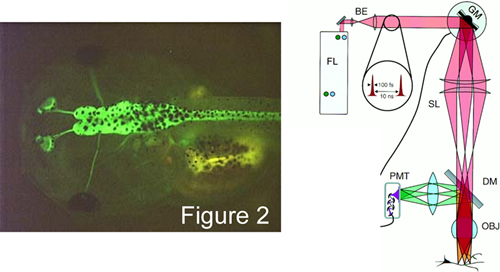
Given this advance, another BME PhD student, Ivy Dick in the laboratory is making transgenic tadpoles that selectively express TNL-15 in the brain (Figure 2, left). Two photon imaging of such animals (Figure 2, right) promise exciting experiments aimed at understanding how visual and auditory stimuli are encoded as neural activity within the living brain. These and like advances now seem possible with the advent of genetically encoded sensors of various bio-signals.